Abstract
Malaria is a major health issue worldwide, and its diagnosis requires scalable solutions that can work effectively with low-cost microscopes (LCM). Deep learning-based methods have shown success in computer-aided diagnosis from microscopic images. However, these methods need annotated images that show cells affected by malaria parasites and their life stages. Annotating images from LCM significantly increases the burden on medical experts compared to annotating images from high-cost microscopes (HCM). For this reason, a practical solution would be trained on HCM images which should generalize well on LCM images during testing. While earlier methods adopted a multi-stage learning process, they did not offer an end-to-end approach. In this work, we present an end-to-end learning framework, named CodaMal (COntrastive Domain Adpation for MALaria). In order to bridge the gap between HCM (training) and LCM (testing), we propose a domain adaptive contrastive loss. It reduces the domain shift by promoting similarity between the representations of HCM and its corresponding LCM image, without imposing an additional annotation burden. In addition, the training objective includes standard object detection losses, ensuring the accurate detection of malaria parasites. On the publicly available large-scale M5-dataset, our proposed method shows a significant improvement of 44% over the state-of-the-art methods in terms of the mean average precision metric (mAP).
Method
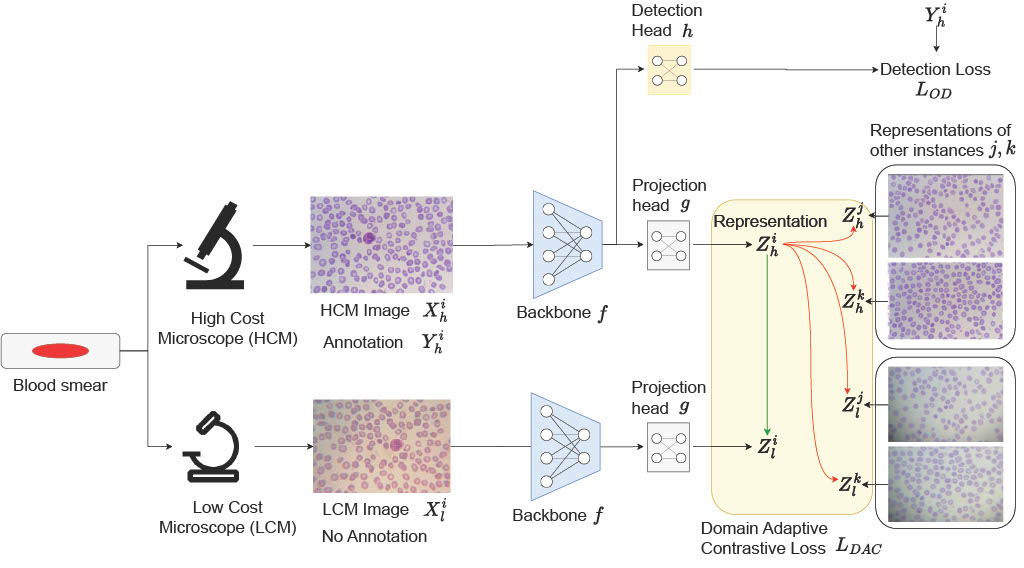
Schematic diagram of our end-to-end method for malaria detection. (a) During training, an HCM image (Xih) and its annotations (Yih) are paired with the corresponding unannotated LCM image (Xil) that is captured from the same field-of-view, instance i. The HCM image (Xih) is utilized to train the backbone (f) and its detection head (h) using the standard object detection losses (LOD). For domain adaptation, both Xih and Xil are processed through the same backbone (f) and non-linear projection head (g). The HCM representation, Zih, is encouraged to maximize similarity with the corresponding LCM instance (Zil) (indicated by the green arrow), while minimizing similarity with the representations of other LCM and HCM image instances (j,k) i.e. instances captured from different FOV or different blood smear (highlighted by the red arrow). (b) Once trained, the model receives LCM images and predicts the location and life-stage of the malaria parasite as its final output.
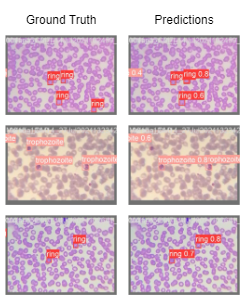
Results
Our method, CodaMal, significantly outperforms prior best results by 42.93% and 44.08% on 1000× and 400× of FOV in terms of mAP and sets a new state-of-art. Also, it is worth noting that our model utilizes only 21.2M parameters which is slightly less than ResNet50 (23M) used by prior work. Apart, from mAP metric, our method achieves a precision of 0.8761 and Recall of 0.4571, on 1000× magnification FOV setting.
1000x
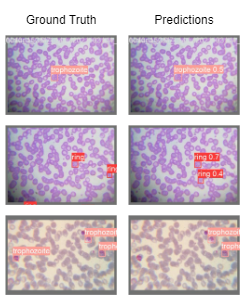
400x
100x

1000x

400x

100x

| Method | End-to-End Training? | Number of Params (M) | Inf. time (ms) | HCM → LCM | |
|---|---|---|---|---|---|
| 1000x | 400x | ||||
| Chen et al.. CVPR'18 | ✓ | 43.7 | 184 | 17.6 | 21.5 |
| Saito et al. CVPR'19 | ✓ | 43.7 | 184 | 24.8 | 21.4 |
| Xu et al. CVPR'20 | ✓ | 43.7 | 184 | 15.5 | 21.6 |
| Faster RCNN | ✓ | 43.7 | 184 | 17.1 | 26.7 |
| Faster RCNN + Synthetic LCM Tuning | ✗ | 43.7 | 184 | 33.3 | 31.8 |
| Faster RCNN + Synthetic LCM Tuning + Ranking Loss | ✗ | 43.7 | 184 | 35.7 | 32.4 |
| Faster RCNN + Synthetic LCM Tuning + Triplet Loss | ✗ | 43.7 | 184 | 37.2 | 32.2 |
| Sultani et al. CVPR'22 | ✓ | 43.7 | 184 | 37.5 | 33.8 |
| CodaMal (Ours) | ✓ | 21.2 | 8.9 | 53.6 | 48.7 |
The performance metric is mAP @ 0.5 IoU. 1000× and 400× represent the magnification FOV of LCM images. Highlighted red shows the best results and blue shows second best results.
Conclusion
In conclusion, our study presents a novel end-to-end train-able method for the more practical setting of deep learning learning-based malaria parasite detection: training on HCM and testing on LCM images. The incorporation of the domain adaptive contrastive loss bridges the domain gap by pro-moting invariance between HCM and LCM domains. Our method, CodaMal, substantially outperforms the prior best methods up to 44% and sets a new state-of-the-art. We are planning to open-source our implementation code upon the acceptance of this work. These findings not only highlight the potential of our method to enhance the early and accurate diagnosis of malaria but also emphasize the importance of domain adaptation in improving the generalization capabilities of deep learning-based models for microscopy applications. Future research directions may explore the application of our method to other microscopy-based diagnostic tasks, further refining the method and assessing its impact on a broader range of diseases and detection scenarios.
BibTeX
@INPROCEEDINGS{dave2024codamal,
author={Dave, Ishan Rajendrakumar and Blegiers, Tristan de and Chen, Chen and Shah, Mubarak},
booktitle={2024 IEEE International Conference on Image Processing (ICIP)},
title={Codamal: Contrastive Domain Adaptation for Malaria Detection in Low-Cost Microscopes},
year={2024},
pages={3848-3853}}